Robust Bayesian Graphical Regression Models for Assessing Tumor Heterogeneity in Proteomic Networks
Abstract
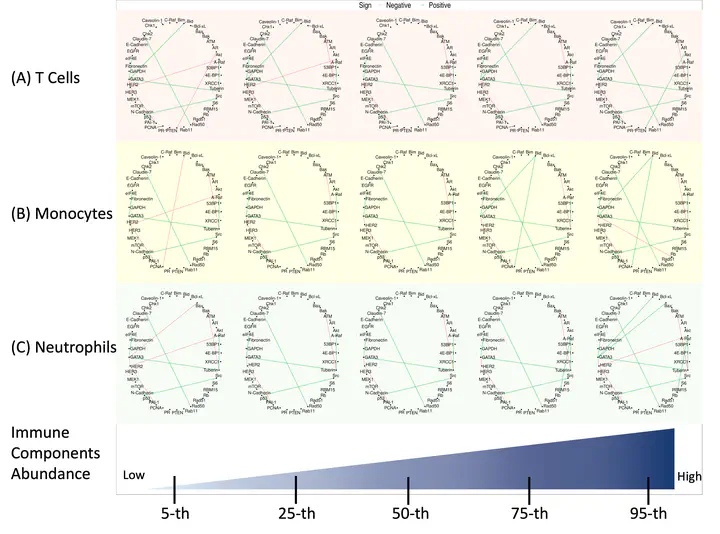
Graphical models are powerful tools to investigate complex dependency structures in high-throughput datasets. However, most existing graphical models make one of the two canonical assumptions of (i) a homogeneous graph with a common network for all subjects, or (ii) an assumption of normality especially in the context of Gaussian graphical models. Both assumptions are restrictive and can fail to hold in certain applications such as proteomic networks in cancer. To this end, we propose an approach termed robust Bayesian graphical regression (rBGR) to estimate heterogeneous graphs for non-normally distributed data. rBGR is a flexible framework that accommodates non-normality through random marginal transformations and constructs covariate-dependent graphs to accommodate heterogeneity through graphical regression techniques. We formulate a new characterization of edge dependencies in such models called conditional sign independence with covariates along with an efficient posterior sampling algorithm. In simulation studies, we demonstrate that rBGR outperforms existing graphical regression models for data generated under various levels of non-normality in both edge and covariate selection. We use rBGR to assess proteomic networks across two cancers, lung and ovarian, to systematically investigate the effects of immunogenic heterogeneity within tumors. Our analyses reveal several important protein-protein interactions that are differentially impacted by the immune cell abundance; some corroborate existing biological knowledge whereas others are novel findings. Our accompanying code and data are available here.